Platinum-E (Plat-E) Retroviral Packaging Cell Line
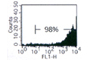
- Higher retroviral yields: average titer 106 to 107 infectious units/mL with transient transfection
- Longer stability: up to 4 months in the presence of drug selection
- Produces ecotropic retrovirus, which can only readily infect mouse or rat cells
NOTE: Platinum Retroviral Packaging Cells are available for sale to academic, government and non-profit research laboratories. All other purchasers require a commercial license for all fields including research use. Please contact our Business Development department for license information.